
A student studies in the open spaces of the Genome Sciences Building at the University of North Carolina at Chapel Hill.
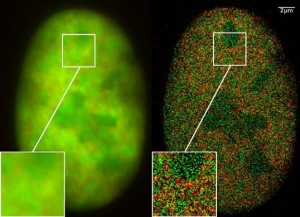
Nucleus of a bone cancer cell with traditional microscopy (left) and super-resolution microscopy (right). Histones are red (RFP-H2A) and chromatin remodeling proteins are green (GFP-Snf2H). Two color localization microscopy reveals single molecule density in a wide field of view using conventional fluorescent proteins.
Since many types of human disease, including a variety of cancers, can be considered to arise from a loss of cellular identity, identifying the mechanisms that regulate proper gene expression programs is important for many aspects of human health. Furthermore, a lot of human variation occurs within regulatory elements and is thought to be responsible for the varying disease-associated risks observed across a population. We seek to identify the cancers that result from perturbed chromosome structure and understand the underlying disease mechanisms in order to inform more effective and targeted therapies for patients. To gain insight into the role of genome organization in healthy cells and cancer cells, we are developing models in a variety of mammalian cell lines to study altered chromosome structures and gene expression programs.
