
A student studies in the open spaces of the Genome Sciences Building at the University of North Carolina at Chapel Hill.
How are DNA loops regulated during development to generate and maintain the diversity of cell types found in an organism?
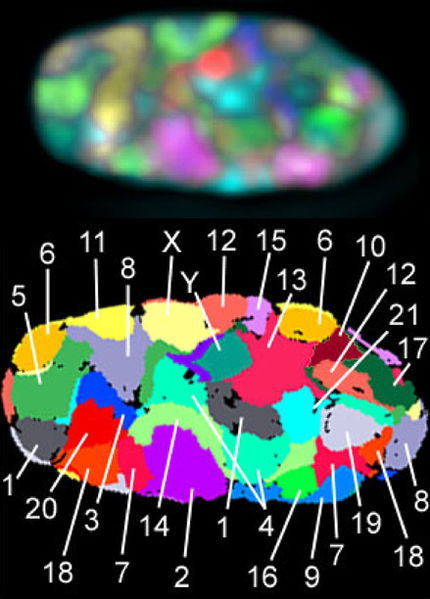
Three-dimensional position of chromosomes in a nucleus. Fluorescence in situ hybridization of all 24 human chromosomes in a human male fibroblast cell. Deconvoluted image (top) from 8 channel wide-field microscopy and false color representation (bottom) of all chromosome territories.
We seek to understand the molecular mechanisms by which cells acquire their identity during development and how dynamic changes in cell identity occur during differentiation. How does a cell faithfully maintain the proper expression of thousands of genes that determine its identity? How is a change brought about when a cell needs to differentiate or respond to an environmental signal? The answers to these questions lie in understanding the dynamic nature of chromatin and determining what instructions are encoded within DNA versus epigenetically regulated. We are investigating the genetic and epigenetic mechanisms that are responsible for control of cellular identity. We use embryonic stem cells as a model system to address this question since they are pluripotent and can be directed to differentiate into the many other cell types of an organism.
